Published online Nov 25, 2014. doi: 10.5495/wjcid.v4.i4.41
Revised: July 24, 2014
Accepted: October 1, 2014
Published online: November 25, 2014
Processing time: 164 Days and 1.3 Hours
Recently, growing evidences show that the combination of epigenetic and genetic abnormalities contribute together to the development of liver diseases. DNA methylation is a very important epigenetic mechanism in human beings. It refers to addition of the methyl groups to DNA and mainly occurs at cytosine adjacent to guanine. DNA methylation is prevalent across human genome and is essential for the normal human development, while its dysfunction is associated with many human diseases. A deep understanding of DNA methylation may provide us deep insight into the origination of liver diseases. Also, it may provide us new tools for diseases diagnosis and prognosis prediction. This review summarized recent progress of DNA methylation study and provided an overview of DNA methylation and liver diseases. Meanwhile, the association between DNA methylation and liver diseases including hepatocellular carcinoma, liver fibrosis, nonalcoholic steatohepatitis and liver failure were extensively discussed. Finally, we discussed the potential of DNA methylation therapeutics for liver diseases and the value of DNA methylation as biomarkers for liver diseases diagnosis and prognosis prediction. This review aimed to provide the emerging DNA methylation information about liver diseases. It might provide essential information for managing and care of these patients.
Core tip: This review summarized recent progress of DNA methylation study and provided an overview of DNA methylation and liver diseases. The association between DNA methylation and liver diseases including hepatocellular carcinoma, liver fibrosis, nonalcoholic steatohepatitis or liver failure were extensively discussed. We also discussed the potential of DNA methylation as biomarkers and therapeutic targets for liver diseases. This review aimed to provide the emerging DNA methylation information about liver diseases. It might provide essential information for managing and care of these patients.
- Citation: Gao S, Wang K. DNA methylation in liver diseases. World J Clin Infect Dis 2014; 4(4): 41-49
- URL: https://www.wjgnet.com/2220-3176/full/v4/i4/41.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v4.i4.41
Because of the high prevalence, liver diseases have been studied systematically during the past few decades. Many studies focus on genetic defects[1] and genome-wide association studies do provide us great information about the pathogenesis of liver diseases[2]. However, many questions which cannot be totally illustrated by genetic mechanism still exist, which lead researchers to initiate the study of epigenetic variation. Recent studies showed that the combination of genetic and epigenetic variants contributed together to the susceptibility and progression of liver diseases[3-5]. Epigenetics refers to the heritable changes of gene expression without changes in gene sequence[6]. DNA methylation is a very important epigenetic mechanism in human and distribute widely across human genome. It is of crucial important for normal development, genomic imprinting as well as inactivation of X-chromosome[7-9]. Meanwhile, aberrant DNA methylation usually associates with many human diseases[10]. The goal of this article is to review the studies associated with DNA methylation and liver diseases. Finally, we look into the future prospect that DNA methylation may bring to the detection and treatment of liver diseases.
DNA methylation which refers to addition of the methyl groups to DNA is firstly introduced in 1970s[11,12]. In invertebrates and fungi, DNA methylation only presents in small proportion of genome and varies among different clades[13,14]. In vertebrate genome, it presents in almost everywhere across the genome. Mainly, DNA methylation occurs at cytosine adjacent to guanine (CpG dinucleotides)[15]. In human genome, The CpG dinucleotides are very rare (approximately 1%). They are nonuniformly distributed and tend to cluster together to form CpG island (CGI). CGI refers to a 200-bp region in DNA which is characterized by high G+C content (more than 50%) and high observed CpG/expected CpG ratio (at least 0.6)[16]. Previous studies showed that CGIs existed in more than half of the genes in vertebrate genomes. Until now, the exact role of gene methylation in gene regulation remains largely unclear[17].
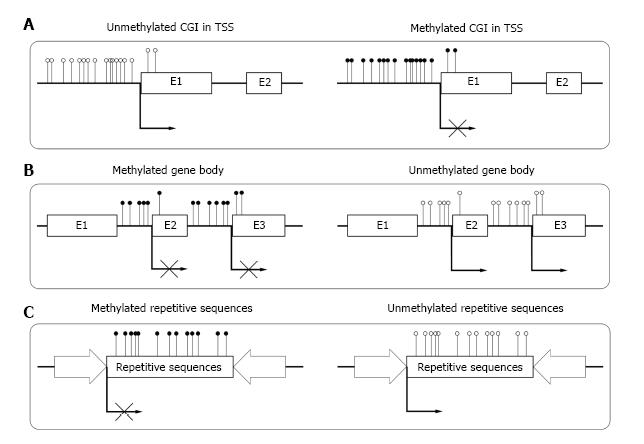
Until now, most of the studies on DNA methylation focus on CGIs in the transcriptional start sites (TSSs) of genes. In human genome, about 60% of gene TSSs contain CGIs and usually remain unmethylated in normal cells. Methylation of these CGIs often result in long-term stabilization of transcriptional silencing and loss of gene function both physically and pathologically[18] (Figure 1A). CpG island shore is defined as lower CpG density region which is close (approximately 2 kb) to the CGI. Recent studies show that most tissue specific methylation occurs at CpG island shores[19,20]. Aberrant DNA methylation at CpG island shores correlate even more strongly with gene expression than CGI[21].
There are about 40% of human genes which do not contain bona fide CGI at their TSSs[16]. Compared with genes that contained CGIs, the role of methylation in genes without CGIs at the TSSs has not been well demonstrated. More studies still need to be performed on genes without CGIs. Studies revealed that maspin gene had a promoter that did not reach the criteria for CGI and hypermethylation of this promoter was strongly correlated with its tissue specific expression[22]. However, MAGE gene was found to be unregulated by methylation in the promoters which do not satisfy CGIs.
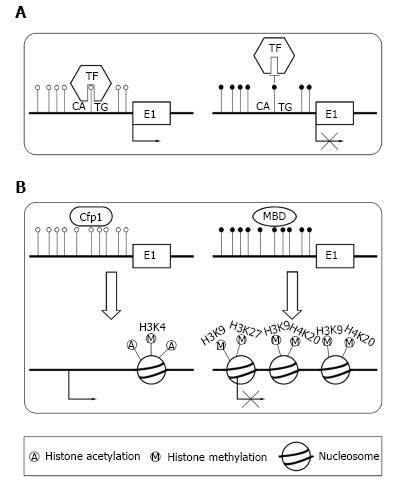
There are two primary means by which DNA methylation in TSSs repress transcription. The transcription factors[23] control gene expression level. DNA methylation can directly preclude the transcription factors binding to its normal sites[24,25] (Figure 2A). For example, transcription factor YY1 which is essential for the imprinting of Peg3 gene can bind to PEG3-DMR sequence in the first intron[24]. In vivo, the methylation of PEG3-DMR sequence precludes the binding of YY1, which result in the repression of maternal allele. In paternal allele, YY1 can effectively bind to the unmethylated PEG3-DMR sequence. Alternatively, DNA methylation can recruit specific proteins and induce a repressive chromatin structure[9] (Figure 2B). In normal condition, unmethylated CGIs can recruit CpG binding proteins, which form a structure suitable for transcription[26]. When CGIs are methylated, they can recruit methyl-CpG-binding domain (MBD) proteins[14,27]. Then, MBD proteins could recruit the histone modifying as well as chromatin remodeling complex to the methylated positions, which result in transcriptional silencing by repressing the transcriptional permissiveness of chromatins.
Although CGIs also exist within gene bodies[28], most gene bodies are CpG-poor and extensively methylated. Studies showed that high level of gene body methylation was positively correlated with transcription, which meant it might associate with gene activation[29,30]. Zilberman et al[31] found that the methylation of gene body could increase elongation efficiency and prevent spurious initiations of transcription (Figure 1B). Shukla et al[32] illustrated that methylation between exons and introns was involved in regulating splicing[33]. Other studies reported that the methylation in gene body could be an important mechanism for managing promoter usage[34]. The high methylation level in gene body was essential for the elongation of a transcript.
Repetitive elements comprise up to 45% of human genome[35], which mainly consist of interspersed repeats and tandem repeats. In normal somatic cells, repetitive sequences of genome are highly methylated. The deeply methylated condition is essential for the stability of chromosome and normal gene expression[36] (Figure 1C). Demethylation of repetitive sequences in genome may result in different kinds of diseases[37,38].
DNA methylation is an important way to store hereditary information. Although it does not change gene sequence, it can propagate the methylation mark during cell divisions[39]. The DNA methylation inheritance process is catalyzed by DNA methyltransferase (DNMT) enzyme family. Manly, there are five members in DNMT enzyme family, DNMT1, DNMT2, DNMT3a, DNMT3b and DNMT3L. DNMT1, DNMT3a, DNMT3b serve as methyltransferase. Each of the three DNMTs is essential for normal human development[7,40]. Studies revealed that loss of methylation resulted from the inactivation of DNMTs could result in apoptosis of somatic cell[41] and cancer cells[42]. However, it showed that DNMTs were not essential for the survival of embryonic stem cells[43].
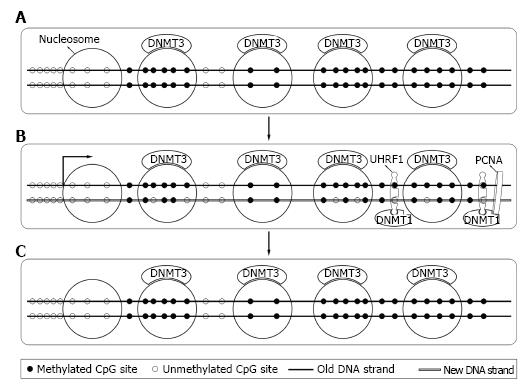
Bestor et al[44] firstly cloned DNMT1 in 1988 from mouse cells. Later studies revealed that DNMT1 expressed mostly at S phase of cell cycle[45] and mainly acted as maintenance DNMT. Interacting with the DNA polymerase processing factor proliferating cell nuclear antigen and ubiquitin-like plant homeodomain and RiNG finger domain containing protein 1 (UHRF1), DNMT1 methylated the hemimethylated sites during DNA semi-reserved replication[46,47]. Soon after replication, DNMT3a and DNMT3b bound to methylated DNA and corrected methylation sites missed by DNMT1 and completed the process[48,49] (Figure 3). DNMT1 was essential for both normal somatic cells and cancer cells and a knockout of DNMT1 could cause their death[41,42].
After the cloning of DNMT1, studies found that embryonic stem cells could still methylate retroviral DNA de novo even without DNMT1[50]. DNMT3a and DNMT3b were found in later studies[40]. They were regarded as de novo DNMT and functioned to set up normal methylation pattern during embryonic development. They were abundant in embryonic stem cell and less expressed in differentiated cells[51]. Other DNMTs like DNMT3L possessed no methylation catalytical activities. But Bourc’his et al[52] found that DNMT3L was crucial for establishment of maternal genomic imprinting.
In hepatocellular carcinoma (HCC), DNA methylation is characterized by a genome wide hypomethylation and a site specific hypermethylation[53]. Until now, many studies for presenting the DNA methylation patterns in HCC have been published.
Compared with normal liver tissue, DNA methylation in HCC is characterized by global hypomethylation. The hypomethylation of intergenic areas, repetitive DNA sequences[54], introns[55] and promoter CGI of specific oncogene[56] are responsible for the global hypomethylation. Global hypomethylation mainly result in chromosomal instability, loss of genomic imprinting[57,58] and reactivation of transposable elements, which may contribute to the development of cancer.
Previous studies revealed that the demethylation of chromosome 1 heterochromatin DNA was associated with the q-arm copy gain[59] in HCC. Also, a number of hypomethylated tumor-promoting genes, including HPA[60], MAT2A[61], VIM[62] and SNCG[63] have been identified in primary human HCC.
In tumor suppressor gene, the hypermethylation of CGIs in TSSs result in the loss of gene function, which is crucial for the origin of cancer[64]. The inactivation of tumor suppressor genes caused by hypermethylation of CGI in TSS exist in almost every type of human cancers[65]. Hypermethylation may affect the process of cell cycle regulation, DNA repair, angiogenesis, programmed cell death and tumor cell invasion. The genes silenced by hypermethylation in human cancers are often those who are essential for the maintenance of stem cell characteristics and/or the maturation of adult cells during cell renewal[65,66]. Silencing of these genes may result in the initiation of tumors through distribution of abnormal stem cells and/or abnormal of normal cell differentiation.
Until now, many tumor suppressor genes have been identified to be hypermethylated in HCC. Table 1 presents a group of frequently methylated genes in HCC.
| Gene | Location | Function | Methylation frequency | Ref. |
| GSTP1 | 11q13.2 | Detoxification | 41%-58% | [85-87] |
| SOCS1 | 16p13.13 | Cytokine inhibitor | 60% | [88] |
| RASSF1A | 3p21.3 | Apoptosis | 54%-95% | [89,90] |
| E-Cadherin | 16q22.1 | Cell adhesion | 33%-67% | [91,92] |
| APC | 5q22.2 | Signal transduction | 46% | [93] |
| p16 | 9q21.3 | CDK inhibitor | 17%-83% | [94,95] |
| SFRP1 | 8p11.21 | Signal transduction | 59.50% | [96] |
| WIF-1 | 12q14.3 | Signal transduction | 61.90% | [97] |
| MGMT | 10q26 | DNA repair | 22%-39% | [98,99] |
| TFPI2 | 7q21.3 | Protease inhibitor | 46.50% | [100] |
In liver fibrosis, aberrant DNA methylation has been studied for a few years. Until now, a number of aberrantly methylated genes have already been recognized. Through direct or indirect examination methods (treated with demethylating agents such as 5-aza-2’-deoxycytidine), these genes were identified to be aberrantly methylated. In activated hepatic stellate cell (HSC), transcriptional repression of some genes was indentified to be due to promoter hypermethylation of them.
Until now, genome-wide studies of DNA methylation associated with HSC activation were limited. Aberrant methylation associated with HSC activation had been reported at specific loci such as the phosphatase and tensin homologue (PTEN) and patched1 (PTCH1) genes. These genes were aberrantly methylated in the myofibroblast and associated with the decreased of gene expression[67,68]. Our previous study revealed that aberrant promoter methylation of PPAR gamma gene was significantly associated with liver fibrosis in patients with chronic hepatitis B[69]. Other genes like Ras GTPase activating-like protein 1 (RASAL1) gene were also found to be aberrantly hypermethylated in liver fibrosis[70].
So far, the relationship between DNA methylation and metabolic diseases was firmly established. Ahrens et al[71] used array-based DNA methylation and mRNA expression profiling to analyze the liver tissues from patients with non-alcoholic fatty liver disease (NAFLD) (n = 45) and health controls (n = 18). Aberrant methylation and decreased mRNA expression were seen for nine genes, which included genes for key enzymes in intermediate metabolism (ACLY, PC and PLCG1) and insulin or insulin-like signaling (IGFBP2, IGF1 and PRKCE)[71]. Studies showed that supplementation of diets lack of methyl donors could induce DNA hypomethylation and the development of steatosis in mice. However, supplementation of diets with methyl donors could prevent the development of NAFLD, suggesting that differences in the DNA methylation status might be a potential factor for individual susceptibilities to hepatic steatosis[72,73]. The supplementation of the maternal diet with methyl donors could induce aberrant methylation in adulthood and protect offspring from suffering obesity[74].
Recent studies found that the aberrant methylation of several genes might participate in the development of liver failure. The aberrant promoter methylation of some anti-inflammatory genes might result in the down-regulate gene expression and inhibit their protective role in liver injury. Our previous study found that glutathione-S-transferase P1 (GSTP1) promoter hypermethylation occurred in patients with acute on chronic hepatitis B liver failure (ACHBLF) which might facilitate oxidative stress associated liver damage[75]. A study performed by Fan et al[76] showed that hypomethylation of IFN-γ gene promoter in peripheral blood mononuclear cells might be associated with the onset of ACHBLF. Qi et al[77] found that the aberrant hypermethylation of glutathione-S-transferase P1 (GSTM3) gene occurred in ACHBLF, which was correlated with their disease severity.
The development of liver diseases is a multifactorial process characterized by the combination and integration of a multitude of alterations including genetic and epigenetic changes. In the past decades, there were exponential increases in the interest and progress of DNA methylation. Studies already revealed the potential role that DNA methylation played in the normal human development and initiation of diseases. DNA methylation-based biomarkers were proposed for disease risk assessment[78], early detection[79,80], prognostic prediction[81] and treatment outcome prediction of liver diseases[82]. Meanwhile, there was hope for developing therapeutic agents to manipulate aberrant DNA methylation patterns and to treat malignancies[6]. In 1970s, Constantinides et al[83] reported 5-azacytidine had remarkable effects on differentiated states of cells. In 2005, Brueckner et al[84] reported the drug RG101 could also reactivate tumor suppressor gene by inhibiting human DNA methyltransferase. Therefore, combined genetic and epigenetic information may help clinicians to prevent liver diseases developing in at-risk individuals and from passing on unhealthy DNA methylation characteristics to offsprings.
P- Reviewer: Andrisani OM, Chen LY S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Mann DA. Epigenetics in liver disease. Hepatology. 2014;60:1418-1425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 2. | Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362-9367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3051] [Cited by in RCA: 3035] [Article Influence: 189.7] [Reference Citation Analysis (0)] |
| 3. | Timmer MR, Beuers U, Fockens P, Ponsioen CY, Rauws EA, Wang KK, Krishnadath KK. Genetic and epigenetic abnormalities in primary sclerosing cholangitis-associated cholangiocarcinoma. Inflamm Bowel Dis. 2013;19:1789-1797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Ozen C, Yildiz G, Dagcan AT, Cevik D, Ors A, Keles U, Topel H, Ozturk M. Genetics and epigenetics of liver cancer. N Biotechnol. 2013;30:381-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 5. | Zimmer V, Lammert F. Genetics and epigenetics in the fibrogenic evolution of chronic liver diseases. Best Pract Res Clin Gastroenterol. 2011;25:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | American Association for Cancer Research Human Epigenome Task Force, European Union, Network of Excellence, Scientific Advisory Board. Moving AHEAD with an international human epigenome project. Nature. 2008;454:711-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 7. | Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915-926. [PubMed] |
| 8. | Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1628] [Cited by in RCA: 1522] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 9. | Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4367] [Cited by in RCA: 4213] [Article Influence: 191.5] [Reference Citation Analysis (0)] |
| 10. | Ray K. NAFLD: Profiling NAFLD--liver gene expression and DNA methylation patterns to characterize disease severity. Nat Rev Gastroenterol Hepatol. 2013;10:565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 11. | Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9-25. [PubMed] |
| 12. | Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226-232. [PubMed] |
| 13. | Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3811] [Cited by in RCA: 4157] [Article Influence: 319.8] [Reference Citation Analysis (0)] |
| 14. | Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2107] [Cited by in RCA: 2194] [Article Influence: 156.7] [Reference Citation Analysis (0)] |
| 15. | Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3346] [Cited by in RCA: 3330] [Article Influence: 208.1] [Reference Citation Analysis (0)] |
| 16. | Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740-3745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1022] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 17. | Schübeler D. Molecular biology. Epigenetic islands in a genetic ocean. Science. 2012;338:756-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 447] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 19. | Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1742] [Cited by in RCA: 1642] [Article Influence: 102.6] [Reference Citation Analysis (0)] |
| 20. | Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350-1353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 912] [Cited by in RCA: 873] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 21. | Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338-342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 480] [Cited by in RCA: 492] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 22. | Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 326] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 23. | Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. [PubMed] |
| 24. | Kim J, Kollhoff A, Bergmann A, Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum Mol Genet. 2003;12:233-245. [PubMed] |
| 25. | Perini G, Diolaiti D, Porro A, Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc Natl Acad Sci USA. 2005;102:12117-12122. [PubMed] |
| 26. | Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 474] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 27. | Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer. 2008;98:1881-1885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 117] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34-37. [PubMed] |
| 29. | Wolf SF, Jolly DJ, Lunnen KD, Friedmann T, Migeon BR. Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X chromosome: implications for X-chromosome inactivation. Proc Natl Acad Sci USA. 1984;81:2806-2810. [PubMed] |
| 30. | Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141-1143. [PubMed] |
| 31. | Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61-69. [PubMed] |
| 32. | Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 718] [Cited by in RCA: 751] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 33. | Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 765] [Cited by in RCA: 768] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 34. | Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253-257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1253] [Cited by in RCA: 1258] [Article Influence: 83.9] [Reference Citation Analysis (0)] |
| 35. | Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16054] [Cited by in RCA: 14995] [Article Influence: 624.8] [Reference Citation Analysis (0)] |
| 36. | Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286-298. [PubMed] |
| 37. | Moarefi AH, Chédin F. ICF syndrome mutations cause a broad spectrum of biochemical defects in DNMT3B-mediated de novo DNA methylation. J Mol Biol. 2011;409:758-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 38. | Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823-6836. [PubMed] |
| 39. | Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 243] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 40. | Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247-257. [PubMed] |
| 41. | Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 528] [Cited by in RCA: 522] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 42. | Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391-396. [PubMed] |
| 43. | Woodward JJ, Blair R. Redox modulation of N-methyl-D-aspartate-stimulated neurotransmitter release from rat brain slices. J Neurochem. 1991;57:2059-2064. [PubMed] |
| 44. | Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971-983. [PubMed] |
| 45. | Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108-2113. [PubMed] |
| 46. | Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996-2000. [PubMed] |
| 47. | Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760-1764. [PubMed] |
| 48. | Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 555] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 49. | Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet. 2011;7:e1001286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 50. | Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195-3205. [PubMed] |
| 51. | Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16 Spec No 1:R50-R59. [PubMed] |
| 52. | Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536-2539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1036] [Cited by in RCA: 974] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 53. | Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3397] [Cited by in RCA: 3701] [Article Influence: 160.9] [Reference Citation Analysis (0)] |
| 54. | Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187-190. [PubMed] |
| 55. | Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1674] [Cited by in RCA: 1523] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 56. | Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89-92. [PubMed] |
| 57. | Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1012] [Cited by in RCA: 938] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 58. | Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capellà G, Ribas M. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462-9468. [PubMed] |
| 59. | Wong N, Lam WC, Lai PB, Pang E, Lau WY, Johnson PJ. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am J Pathol. 2001;159:465-471. [PubMed] |
| 60. | Xiao Y, Kleeff J, Shi X, Büchler MW, Friess H. Heparanase expression in hepatocellular carcinoma and the cirrhotic liver. Hepatol Res. 2003;26:192-198. [PubMed] |
| 61. | Yang H, Huang ZZ, Zeng Z, Chen C, Selby RR, Lu SC. Role of promoter methylation in increased methionine adenosyltransferase 2A expression in human liver cancer. Am J Physiol Gastrointest Liver Physiol. 2001;280:G184-G190. [PubMed] |
| 62. | Kitamura Y, Shirahata A, Sakuraba K, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y. Aberrant methylation of the Vimentin gene in hepatocellular carcinoma. Anticancer Res. 2011;31:1289-1291. [PubMed] |
| 63. | Zhao W, Liu H, Liu W, Wu Y, Chen W, Jiang B, Zhou Y, Xue R, Luo C, Wang L. Abnormal activation of the synuclein-gamma gene in hepatocellular carcinomas by epigenetic alteration. Int J Oncol. 2006;28:1081-1088. [PubMed] |
| 64. | Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2672] [Cited by in RCA: 2575] [Article Influence: 151.5] [Reference Citation Analysis (0)] |
| 65. | Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-116. [PubMed] |
| 66. | Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808-811. [PubMed] |
| 67. | Bian EB, Huang C, Ma TT, Tao H, Zhang H, Cheng C, Lv XW, Li J. DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell activation and liver fibrogenesis in rats. Toxicol Appl Pharmacol. 2012;264:13-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 68. | Yang JJ, Tao H, Huang C, Shi KH, Ma TT, Bian EB, Zhang L, Liu LP, Hu W, Lv XW. DNA methylation and MeCP2 regulation of PTCH1 expression during rats hepatic fibrosis. Cell Signal. 2013;25:1202-1211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 69. | Zhao Q, Fan YC, Zhao J, Gao S, Zhao ZH, Wang K. DNA methylation patterns of peroxisome proliferator-activated receptor gamma gene associated with liver fibrosis and inflammation in chronic hepatitis B. J Viral Hepat. 2013;20:430-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 70. | Tao H, Huang C, Yang JJ, Ma TT, Bian EB, Zhang L, Lv XW, Jin Y, Li J. MeCP2 controls the expression of RASAL1 in the hepatic fibrosis in rats. Toxicology. 2011;290:327-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 71. | Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 403] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 72. | Pogribny IP, Tryndyak VP, Bagnyukova TV, Melnyk S, Montgomery B, Ross SA, Latendresse JR, Rusyn I, Beland FA. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 129] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 73. | Cordero P, Campion J, Milagro FI, Martínez JA. Dietary supplementation with methyl donor groups could prevent nonalcoholic fatty liver. Hepatology. 2011;53:2151-2152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 74. | Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 733] [Cited by in RCA: 613] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 75. | Li T, Meng QH, Zou ZQ, Fan YC, Long B, Guo YM, Hou W, Zhao J, Li J, Yu HW. Correlation between promoter methylation of glutathione-S-tranferase P1 and oxidative stress in acute-on-chronic hepatitis B liver failure. J Viral Hepat. 2011;18:e226-e231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 76. | Fan XP, Zou ZQ, Long B, Guo YM, Wang SK, Jia DX, Xu AL, Li FC, Fan YC, Wang K. Enhanced demethylation of interferon-γ gene promoter in peripheral blood mononuclear cells is associated with acute-on-chronic hepatitis B liver failure. Tohoku J Exp Med. 2011;224:13-19. [PubMed] |
| 77. | Qi L, Zou ZQ, Wang LY, Gao S, Fan YC, Long B, Guo YM, Xu AL, Han J, Li T. Methylation of the glutathione-S-transferase M3 gene promoter is associated with oxidative stress in acute-on-chronic hepatitis B liver failure. Tohoku J Exp Med. 2012;228:43-51. [PubMed] |
| 78. | Mah WC, Lee CG. DNA methylation: potential biomarker in Hepatocellular Carcinoma. Biomark Res. 2014;2:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 79. | Ji XF, Fan YC, Gao S, Yang Y, Zhang JJ, Wang K. MT1M and MT1G promoter methylation as biomarkers for hepatocellular carcinoma. World J Gastroenterol. 2014;20:4723-4729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 80. | Han LY, Fan YC, Mu NN, Gao S, Li F, Ji XF, Dou CY, Wang K. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a potential biomarker for hepatitis B Virus associated hepatocellular carcinoma. Int J Med Sci. 2014;11:164-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 81. | Honda S, Haruta M, Sugawara W, Sasaki F, Ohira M, Matsunaga T, Yamaoka H, Horie H, Ohnuma N, Nakagawara A. Quantitative analysis of APC promoter methylation in hepatocellular carcinoma and its prognostic implications. Oncol Lett. 2014;7:1683-1688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 82. | Honda S, Haruta M, Sugawara W, Sasaki F, Ohira M, Matsunaga T, Yamaoka H, Horie H, Ohnuma N, Nakagawara A. The methylation status of RASSF1A promoter predicts responsiveness to chemotherapy and eventual cure in hepatoblastoma patients. Int J Cancer. 2008;123:1117-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 83. | Constantinides PG, Jones PA, Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267:364-366. [PubMed] |
| 84. | Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305-6311. [PubMed] |
| 85. | Zhang YJ, Chen Y, Ahsan H, Lunn RM, Chen SY, Lee PH, Chen CJ, Santella RM. Silencing of glutathione S-transferase P1 by promoter hypermethylation and its relationship to environmental chemical carcinogens in hepatocellular carcinoma. Cancer Lett. 2005;221:135-143. [PubMed] |
| 86. | Zhong S, Tang MW, Yeo W, Liu C, Lo YM, Johnson PJ. Silencing of GSTP1 gene by CpG island DNA hypermethylation in HBV-associated hepatocellular carcinomas. Clin Cancer Res. 2002;8:1087-1092. [PubMed] |
| 87. | Anzola M, Cuevas N, López-Martínez M, Saiz A, Burgos JJ, de Pancorbo MM. No association between GSTP1 gene aberrant promoter methylation and prognosis in surgically resected hepatocellular carcinoma patients from the Basque Country (Northern Spain). Liver Int. 2003;23:249-254. [PubMed] |
| 88. | Okochi O, Hibi K, Sakai M, Inoue S, Takeda S, Kaneko T, Nakao A. Methylation-mediated silencing of SOCS-1 gene in hepatocellular carcinoma derived from cirrhosis. Clin Cancer Res. 2003;9:5295-5298. [PubMed] |
| 89. | Schagdarsurengin U, Wilkens L, Steinemann D, Flemming P, Kreipe HH, Pfeifer GP, Schlegelberger B, Dammann R. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22:1866-1871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 145] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 90. | Zhong S, Yeo W, Tang MW, Wong N, Lai PB, Johnson PJ. Intensive hypermethylation of the CpG island of Ras association domain family 1A in hepatitis B virus-associated hepatocellular carcinomas. Clin Cancer Res. 2003;9:3376-3382. [PubMed] |
| 91. | Matsumura T, Makino R, Mitamura K. Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin Cancer Res. 2001;7:594-599. [PubMed] |
| 92. | Kwon GY, Yoo BC, Koh KC, Cho JW, Park WS, Park CK. Promoter methylation of E-cadherin in hepatocellular carcinomas and dysplastic nodules. J Korean Med Sci. 2005;20:242-247. [PubMed] |
| 93. | Jain S, Chang TT, Hamilton JP, Lin SY, Lin YJ, Evans AA, Selaru FM, Lin PW, Chen SH, Block TM. Methylation of the CpG sites only on the sense strand of the APC gene is specific for hepatocellular carcinoma. PLoS One. 2011;6:e26799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 94. | Tannapfel A, Wittekind C. Genes involved in hepatocellular carcinoma: deregulation in cell cycling and apoptosis. Virchows Arch. 2002;440:345-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 95. | Kaneto H, Sasaki S, Yamamoto H, Itoh F, Toyota M, Suzuki H, Ozeki I, Iwata N, Ohmura T, Satoh T. Detection of hypermethylation of the p16(INK4A) gene promoter in chronic hepatitis and cirrhosis associated with hepatitis B or C virus. Gut. 2001;48:372-377. [PubMed] |
| 96. | Nomoto S, Kinoshita T, Kato K, Otani S, Kasuya H, Takeda S, Kanazumi N, Sugimoto H, Nakao A. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer. 2007;97:1260-1265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 97. | Huang L, Li MX, Wang L, Li BK, Chen GH, He LR, Xu L, Yuan YF. Prognostic value of Wnt inhibitory factor-1 expression in hepatocellular carcinoma that is independent of gene methylation. Tumour Biol. 2011;32:233-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 98. | Matsukura S, Soejima H, Nakagawachi T, Yakushiji H, Ogawa A, Fukuhara M, Miyazaki K, Nakabeppu Y, Sekiguchi M, Mukai T. CpG methylation of MGMT and hMLH1 promoter in hepatocellular carcinoma associated with hepatitis viral infection. Br J Cancer. 2003;88:521-529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 99. | Zhang YJ, Chen Y, Ahsan H, Lunn RM, Lee PH, Chen CJ, Santella RM. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relationship to aflatoxin B1-DNA adducts and p53 mutation in hepatocellular carcinoma. Int J Cancer. 2003;103:440-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 100. | Sun FK, Fan YC, Zhao J, Zhang F, Gao S, Zhao ZH, Sun Q, Wang K. Detection of TFPI2 methylation in the serum of hepatocellular carcinoma patients. Dig Dis Sci. 2013;58:1010-1015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |